Welcome to the Yasmin Shamsudin Lab
Our group uses computational modeling and simulations to understand molecular mechanisms and interactions between ligands and proteins that drive binding, inhibition, and catalysis.

Designing drugs and proteins
Computational Enzymology
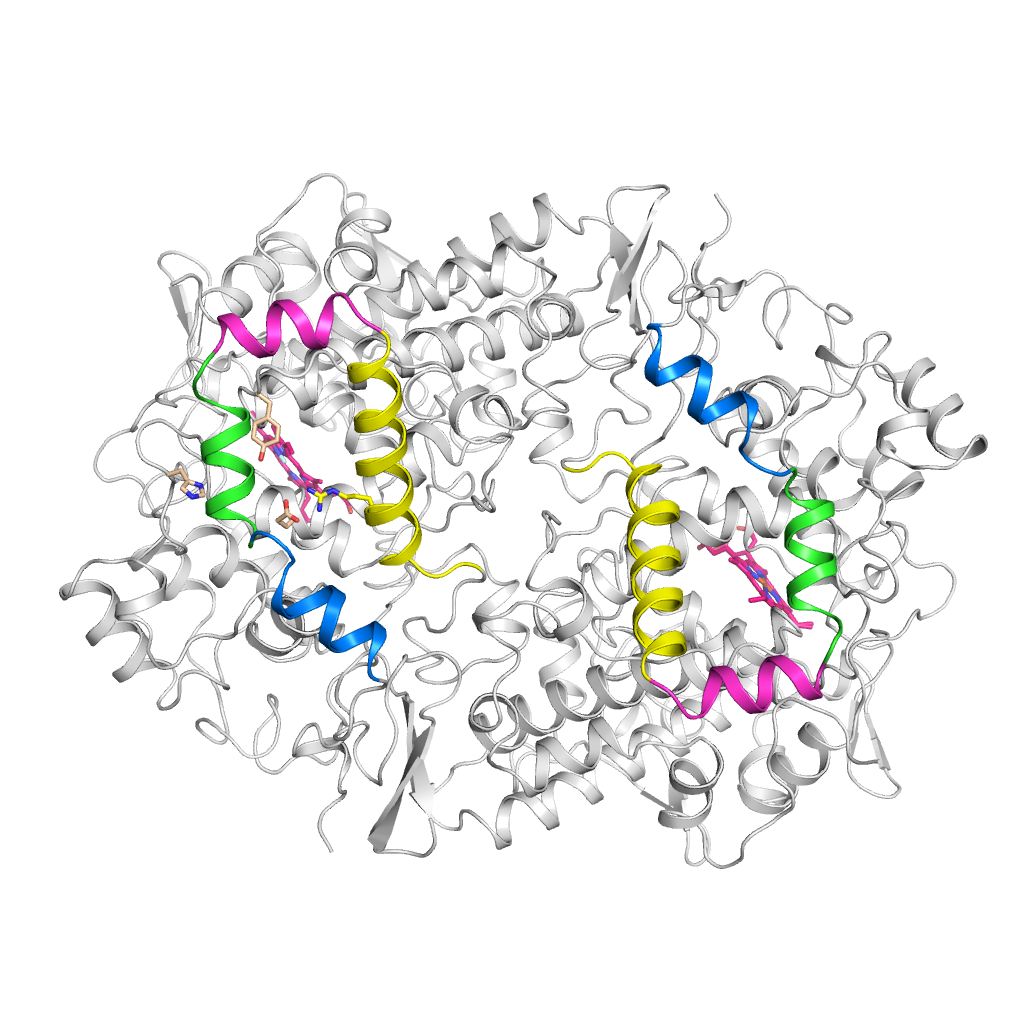
We use molecular modelling and simulations to understand molecular mechanisms regulating inhibition and catalysis in enzymes to guide the design of novel drugs.
Key words: Homology modelling, molecular docking, molecular dynamics simulations, free energy calculations
Electric fields in biomolecules
We developing and validate computational methods for accurately predicting electric fields from molecular modeling and simulations, which could be used to predict catalysis in enzymes.
Key words: Methods development, Enzymatic catalysis
Protein Engineering from computer simulations

We design and engineer functional proteins and enzymes based on predictions from quantum chemical and enhanced sampling molecular dynamics simulations.
Key words: Enhanced sampling molecular dynamics simulations, molecular biology, physical chemistry experiments
Consulting
Need help with your drug design or discovery pipeline? Contact us for more information about how we can help you.
Project Management
Need an experienced project manager for your drug discovery portfolio? Contact us!
Collaborations
Do you have an experimental lab and want additional insight to your results? Contact us to discuss collaborations!