Research
Dedicated to excellence in science
Our group uses computational modeling and simulations to understand molecular mechanisms and interactions between ligands and proteins that drive binding, inhibition, and catalysis.
We work on a range of systems involved in bacterial infections, pain, fever, and inflammation, and Alzheimer’s disease.

Our Projects
Computational Enzymology
Using computational methods like molecular modelling, molecular dynamics simulations, and free energy and electric field calculations, we strive to understand how drugs bind to a range of enzymes, with the objective to create more efficient drugs with less side effects.
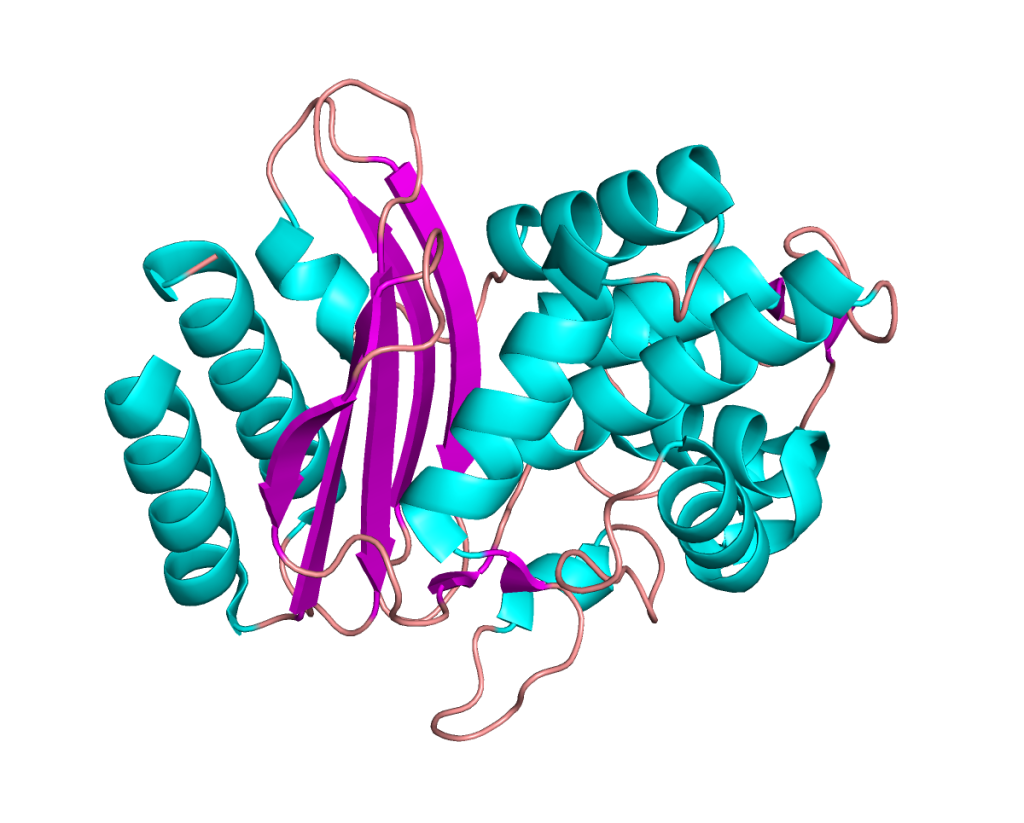
β-lactamase and antibiotics
β-lactamases break down current antibiotics, causing antibiotic resistance.
How do antibiotics and inhibitors bind to these enzymes and which structural features causes enzyme promiscuity? Understanding both factors could lead to new drugs.
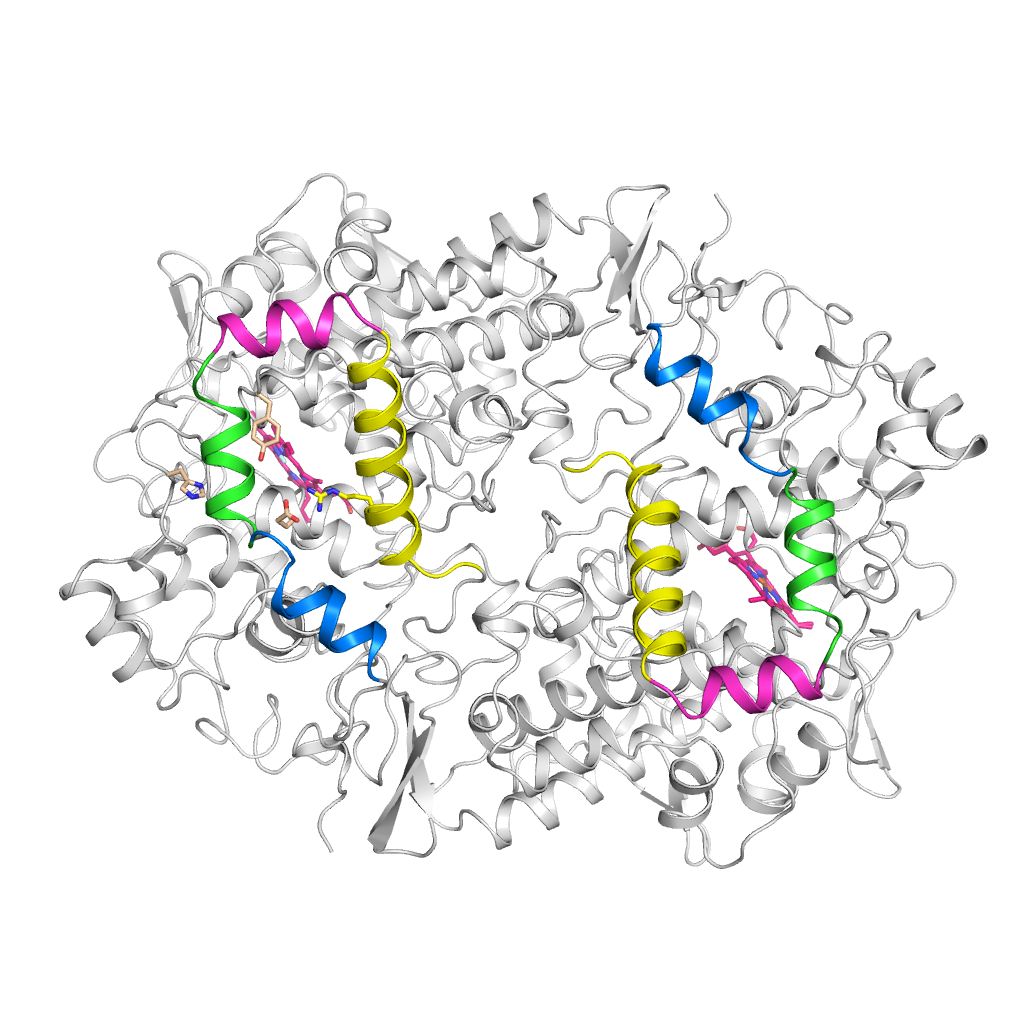
COX-1 & 2 and NSAIDs
Cyclooxygenase (COX) 1 and 2 are the target enzymes of Non-steroidal anti-inflammatory drugs (NSAIDs).
How do NSAIDs bind to these enzymes and which structural features and molecular mechanims causes tight-binding and selectivity towards either isoform?
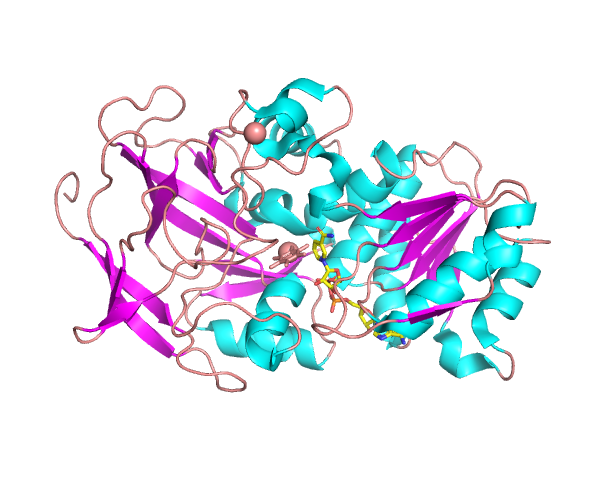
Liver Alcohol Dehydrogenase (LADH)
Liver alcohol dehydrogenases (LADH) convert alcohol to aldehydes.
Is it possible to predict how amino acid substitutions will affect catalytic rates or efficiencies using computational methods?
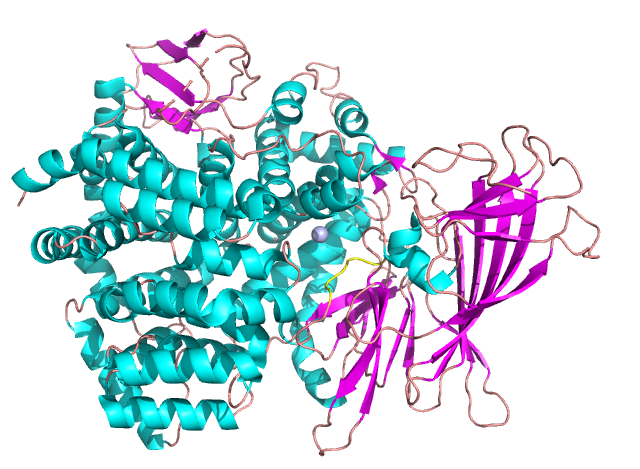
Insulin Regulated Aminopeptidases (IRAP)
Insulin-regulated aminopeptidases (IRAP) are potential targets for treatment of Alzheimers disease.
Understanding the structural features regulating inhibitor binding in IRAP could be the key to designing more efficient inhibitors and future Alzheimers drugs.
Protein Engineering



Collaborate with us!
We are always interested in discussing the needs of experimentalists and theoretical chemists. This enables us to identify synergies and potential for collaborations, to increase the impact of the research, for the benefit of society.